The unknown inner workings that control our genes
This article was originally published in the latest issue of L'Édition (N. 20).
What do the search for new cancer therapies, ripening tomatoes and the prediction of sperm fertility in bulls have in common? It is epigenetics, which is the study of the mechanisms that regulate gene expression without changing the DNA sequence. Because what matters is not always the sequence, but the way in which it is used.
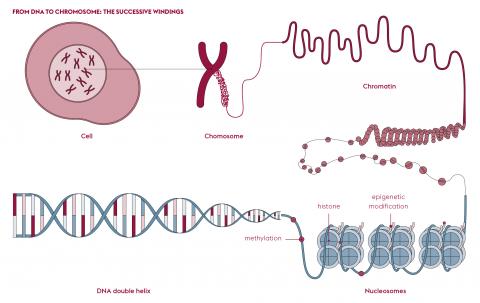
Nestled at the heart of the nucleus of cells, DNA (or deoxyribonucleic acid) contains all the genetic information (or genome) of an individual. This molecule is organised into functional units, the genes, which determine the characteristics of the individual or of a species, much like a cookbook full of recipes. Like a constantly active kitchen brigade, multitudes of molecules are busy in both the nucleus and the cytoplasm of cells. Some are able to recognise the regions of DNA containing genes and initiate the production of messenger ribonucleid acids (mRNAs), and their export to the cytoplasm, while others translate these mRNAs into proteins, which are themselves involved in cellular functions and the life of the individual. But among all the recipes available in the DNA “book”, each cell type chooses which dishes it concocts and in what quantity. What ultimately determines this “menu”?
The main answer comes from DNA compaction. To “fit” the molecule into the nucleus of a cell, several levels of compaction are required. First, DNA is wrapped around proteins, the histones, to form nucleosomes, the basic units of chromatin. Except during cell division, where it is found as chromosomes, chromatin looks like a more or less condensed coil of thread. Epigenetic molecular mechanisms regulate the level of condensation or decondensation of chromatin. These are chemical modifications, applied directly to the histones or to the DNA molecule. These modifications influence gene expression without changing the DNA sequence. Some highly condensed regions are thus packed so tightly that they are unreadable, as if some pages of the book were glued together. To make reading possible, the chromatin must be decondensed. The list of accessible pages therefore differs from one cell type to another and changes over time.
Like food critics, epigenetics researchers at Université Paris-Saclay are interested in the choice and execution of the dishes concocted by the cells: they study more particularly the regulation of epigenetic modifications.
Methylation as a prognosis for cancer
One of the modifications studied is DNA methylation. This is the addition of a methyl group (-CH3), i.e. a carbon atom linked to three hydrogen atoms, on cytosines – nucleic bases that are one of the building blocks of DNA. The cytosines involved are located in a particular DNA sequence environment. This modification usually prevents the expression of nearby genes.
In pioneering work, scientists of the Laboratory for Epigenetics and Environment (LEE) at the National Centre for Research in Human Genomics (CNRGH – Univ. Paris-Saclay, CEA) have focused on this regulation mode. Jörg Tost’s team has discovered that a DNA methylation differential helps to establish a prognosis for breast cancer. Using tumour biopsies, it monitored the epigenetic profile of patients’ tumour cells during chemotherapy. The team found that those who responded well to treatment and survived for several years were those whose the DNA methylation profile of these cells was altered during chemotherapy. The challenge now is to successfully use methylation as an early predictor of long-term survival, starting at the end of drug treatment.
Establishing a full epigenetic assessment
Jörg Tost and his team are also interested in inflammatory diseases. “This research has tremendous potential, because the symptoms observed in inflammatory diseases are almost all related to epigenetics,” says the researcher. “We want to use a simple blood test to find out which epigenetic marks are indicative of the inflammatory state.” For the time being, the technique is only in the early stages of research.
“For the past few years, we have not only been looking at DNA methylation, our approach is more comprehensive.” Chromatin accessibility, changes in histones, coding and non-coding transcriptome, small RNAs: the team scrutinises all levels of epigenetics to understand which molecular changes really have an impact on the functioning of the individual genome. The LEE is now combining its expertise in oncology and immune disease, and is conducting studies in immuno-oncology. The aim is to predict the chances of success of an immunotherapy before it has even begun. “This will be a major topic of study in the coming years. Epigenetics holds the keys to the personalised medicine of the future,” promises Jörg Tost.
Epigenetics: a written record of experience
Another area of study is the environment effects on epigenetic changes. Smoking, exposure to various pollutants or air quality, for example, can modify the epigenetic state of a cell. “The epigenome, which is the set of epigenetic modifications, represents the memory of our exposure. If your mother lived on a farm during her pregnancy and your early childhood, you are much less likely to develop allergies or asthma,” says Jörg Tost. The beneficial bacteria that are in the stables of large animals are the basis of this protection: this is the epigenetics of acquired immunity.
“Everything that happens during gestation can have long-term consequences,” say Hélène Jammes and Anne Gabory, of the Biology of Reproduction, Environment, Epigenetics and Development laboratory (BREED – Univ. Paris-Saclay, UVSQ, INRAE, ENVA). The team studies the link between a change in the maternal environment during gestation and the consequences on the development of the embryo and then the foetus and the fate of the individual post-natally. In particular, it examines the underlying epigenetic mechanisms. “It is important to identify the epigenetic effects on the offspring of all the events occurring during pregnancy,” explains Anne Gabory. “This has an interest for prevention in human medicine but also for livestock farming.” In mice, the maternal metabolic context thus influences foeto-placental development and predisposes to altered responses. When an overweight mouse gives birth to baby mice, the latter are more likely to gain weight in the case of an obesogenic diet. By analysing epigenetic marks present in the blood of the offspring of a litter, the team is trying to predict which mice will be overweight.
However, is the epigenetic profile of parents transmitted to their children and grandchildren, and even beyond? While epigenetic transmission over several generations has been proven in plants, the scientific consensus is not yet established for animals. After fertilisation, almost all epigenetic markers carried by the paternal (from the sperm) and maternal (from the oocyte) genetic heritages are reset. New markers are applied during the embryo’s development, which seems, at first sight, to indicate an absence of epigenetic transmission between parents and children. However, some experimental results show that the erasure wave is not complete. Hélène Jammes explains: “We prefer to talk about epigenetic intergenerational transmission, in parallel with genetic inheritance. It is rather information stored in the cell nucleus during gestation that changes the way cells respond to their environment. Thus, the mice studied are more likely to become obese but are not born overweight.”
Male fertility: bulls and men
Within BREED, scientists are also working on ruminant fertility. “It may seem surprising for the same team to work on different models, but our biological questions and objectives remain the same: to identify the epigenetic mechanisms that contribute to the development of traits and to use them as diagnostic and prognostic tools.” Recent work reports the involvement of epi- genetic information transmitted by the father’s sperm on embryonic development at the time of fertilisation. Genetically characterised bulls, which are selected according to criteria aimed at improving the meat or milk quality of the offspring, are used to inseminate large numbers of cows and obtain large offspring. The fertility of these individuals can then be accurately assessed.
BREED scientists have identified several hundred cytosines whose methylation levels vary in relation to bulls’ fertility. Methylation alterations target genes involved in sperm physiology and embryonic development. “Epigenetics is not a tsunami, but rather a lacework,” says Hélène Jammes. By combining epigenetic data and artificial intelligence, the scientists produced a mathematical model that was able to predict bull fertility at over 70%. This work also provides a better understanding of human male infertility. The bovine model is also being used in the European RUMIGEN project to explore the relationship between paternal transmission of epigenetic information and offspring performance.
Epigenetic diversity at the heart of breeding
The GEroNIMO (Genome and epigenome enabled breeding in monogastrics) project, co-coordinated by Tatiana Zerjal, from the Animal Genetics and Integrative Biology unit (GABI – Univ. Paris-Saclay, INRAE, AgroParisTech) and Frédérique Pitel, is also seeking to understand the extent to which the animal retains memories of environmental changes. “We are looking to see if exposure to stress or other environmental influences leaves traces in DNA methylations. We are studying the possibility of a transmission over several generations, by analysing how changes in the parental environment modify the offspring several generations away.” The chosen study model is a novel one: chickens and their eggs. Identifying the link between epigenetic marks and interesting agricultural traits is also the focus of the GEroNIMO project. This project proposes to analyse DNA and epigenetic marks over fifteen generations of pigs. For farmers, this means selecting their animals on agronomic criteria in the traditional way, and for scientists, monitoring their epigenome over the generations.
“We combine two techniques, GBS (Genotype-by- sequencing) and MeDIP (Methylated-DNAimmunoprecipitation), to scrutinise the genetics and epigenetics of the animals. We analyse thousands of animals, which allows us to study population epigenetics of pigs, laying hens and quails. Our project is focused on the applied consequences,” says Tatiana Zerjal. Here, epigenetics is used for comparisons between free-range and indoor farming, between temperate and tropical climates, or between different breeds of pigs and chickens. Although epigenetic and genetic diversity seem to be linked, a large part of the epigenetic variability is not yet explained. The aim of GEroNIMO is to develop new strategies for selection and conservation of this epigenetic diversity, in order to adapt farming to global changes.
Releasing transposons from tomatoes
In plants, scientists make lines with altered epigenomes to understand how epigenetic memory is passed on between generations. They use the genome editing tool CRISPR-Cas9 or “molecular scissors”. This molecular biology tool makes it possible to cut DNA at a precise location, at the level of specific sequences (CRISPR), using a particular protein (Cas9).
“We are interested in the functional aspect of epigenetics and need these genetic tools associated to bioinformatics to process our large amounts of data. By targeting, for example, the MET1, DML2 or DDM1 genes of the plant, which control DNA methylation, we prevent regulation via this epigenetic mechanism,” explains Nicolas Bouché, researcher at the Jean-Pierre Bourgin Institute (IJPB – Univ. Paris-Saclay, INRAE, AgroParisTech). His team is interested in the model organism Arabidopsis thaliana, the thale cress, a plant little known to the general public but much used by scientists because of its simple and easy-to-study genome. However, the researcher has extended his field of research to a cultivated plant: the tomato. “Epigenetics is of particular importance here. If a tomato plant is prevented from demethylating, the fruit will not ripen.”
The researcher explains: “The epigenome of crop plants is complex. One of the main differences between thale cress and tomatoes is the amount of transposons present in their genome. These are portions of DNA that can move within the genome and modify it by inserting themselves into genes. Their mobility must therefore be very carefully controlled. They represent 20% of the genome in A. thaliana, but 70% in tomatoes. This large quantity of transposons is common to all cultivated species.” At the moment, the role of transposons is not fully understood. They are generally repressed by DNA methylation and remain silent and inactive. In the IJPB greenhouses, plants engineered with CRISPR-Cas9 to decrease their methylation level no longer control their transposons, resulting in radical gene regulation disruptions. The changes observed as a result of these disruptions are used to associate epigenetic regions with a given agronomic trait, such as growth or flowering time. In particular, the team is interested in the epigenetic factors that control plant growth under drought conditions. A major issue in the context of global warming.
Wheat epigenetics
As far as climate change is concerned, there is no doubt that it will have a major impact on crops, especially wheat, which is very sensitive to heat. As part of the 3DWheat project, winner of a 2022 ERC Consolidator grant, Moussa Benhamed’s team, from the Institute of Plant Sciences Paris-Saclay (IPS2 – Univ. Paris-Saclay, CNRS, INRAE, Univ. d’Évry, Univ. Paris Cité) is exploring the epigenetic response of this cereal to high temperatures. When it is too hot, the plant is under stress, which modifies its physiology. This change could be mediated by epigenetic mechanisms.
Moussa Benhamed’s team is particularly interested in histones epigenetic modifications. For example, if one of the histones carries a repression mark, such as methylation of lysine 9 of histone H3, it compacts the neighbouring DNA into a tight coil. On the contrary, if the histone carries an activation mark, such as acetylation of lysine 14 of histone H3, it leaves the DNA around it loose, which allows gene expression. Nearby DNA sequences are thus co-regulated and the genes are expressed at the same time. Sometimes, a histone is doubly marked. The genes at this location are called bivalent and are often involved in the stress response. Moussa Benhamed’s team wants to elucidate the molecular mechanisms at work. “What is the impact of this bivalence? Is the gene response to stress faster? Does this confer a cellular memory?”
Furthermore, as the DNA coil forms loops, it brings genes that are normally distant in the genomic sequence spatially closer together. Under the microscope, the IPS2 scientists observed clumps of an enzyme, RNA polymerase. These agglomerates are real gene expression factories. “These factories connect several loops of DNA together, producing a co-regulation phenomenon between genes that are sometimes thousands of nucleotides apart. Another challenge of the 3DWheat project is to study the role of these epigenetic factories and their dynamics during the response to heat,” concludes Moussa Benhamed.
Epigenetics is a rapidly expanding field that relies on the rapid progress of technologies to explain phenomena that were still unknown a few years ago. Research will play a key role in overcoming future challenges to human health and how human beings adapt to change.
Publications :
- Corem, S., et al. Redistribution of CHH methylation and small interfering RNAs across the genome of tomato ddm1 mutants. The Plant Cell, (2018).
- Costes, V. et al. Predicting male fertility from the sperm methylome: application to 120 bulls with hundreds of artificial insemination records. Clinical epigenetics, vol. 14, 54, (2022).
- Coustham, V., et al. Epigenetics at the interface between environment and DNA: what importance for breeding practices and selection methods? Animal – science proceedings, vol. 13, 5, (2022).
- Fogel, O., et al. Deregulation of microRNA expression in monocytes and CD4+ T lymphocytes from patients with axial spondyloarthritis. Arthritis Res. Ther. 21, 51, (2019).
- Pedersen, C.A., et al. DNA methylation changes in response to neoadjuvant chemotherapy are associated with breast cancer survival. Breast Cancer Res. 24, 43 (2022).
- Safi-Stibler, S., Gabory, A., Epigenetics and the Developmental Origins of Health and Disease: Parental environment signalling to the epigenome, critical time windows and sculpting the adult phenotype. Seminars in Cell & Developmental Biology, vol. 97, (2020).