Developing new antibiotics using gut microbiota
Understanding the enzyme mechanisms of bacteria in gut microbiota to develop new molecules against antibiotic resistance, is one of Olivier Berteau’s pet subjects at the Institute for Food Microbiology for Human Health (MICALIS - Université Paris-Saclay, INRAE, AgroParisTech). This specialist in the biochemistry of microbiota has been awarded an ERC grant for the third time in the Proof of concept category for his ENTRAP project.
The resistance of some bacteria to antibiotics, also known as antibiotic resistance, has become a major public health issue as it is responsible for tens of thousands of deaths per year in Europe. In France alone, antimicrobial resistance is responsible for at least 5,500 deaths per year. The phenomenon of resistance is linked to the misuse of antibiotics in particular. In 2015, the World Health Organization (WHO) warned about the need to optimise the use of antimicrobial drugs, as well as strengthen the research and development of new and effective compounds. Olivier Berteau and his Chemistry and synthetic biology (ChemSyBio) team at the Micalis Institute have been dedicating their work to this area through the study of bacteria in gut microbiota.
A health problem with an alarming scenario
Antibiotic resistance is when an antibiotic treatment is no longer effective against a bacterial infection. Normally, antibiotic treatment neutralises the bacteria responsible for the infection and only a minor amount of resistant bacteria survives in the gut. The repeated administration of antibiotics or their inappropriate use can lead to the prevalence of resistant bacteria. The overuse of antibiotics in particular triggers a ‘selective pressure’ in the body which favours the mutation and multiplication of these bacteria.
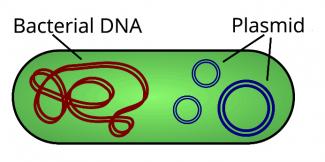
When resistance arises from chromosomal mutations in the bacteria’s DNA, the resistance applies to only one antibiotic or family of antibiotics. However, in 80% of cases, antibiotic resistance is linked to the acquisition of genetic material by ‘conjugation’, i.e. the exchange of plasmids from a donor bacterium to a recipient bacterium. The genetic information carried by the plasmids, such as antibiotic resistant genes, can then be transferred from one bacterium to another.
Source: Normand.cyr
If the plasmid resistance affects several antibiotics, or even several families of antibiotics, then the bacteria are said to be multi-resistant. Resistance transfer via plasmids can occur between different strains and species of bacteria. The accumulation of resistance mechanisms in a single bacterial strain then generates total resistance, i.e. resistance to all available antibiotics, leaving doctors with nowhere to turn to for treatment.
Understanding gut microbiota
Gut microbiota includes all the micro-organisms which live in the small intestine and the colon of a human being, i.e. 1012 to 1014 bacteria, viruses, parasites and nonpathogenic fungi! It is the most important source of micro-organisms in humans and, as yet, an untapped resource of bioactive compounds and antibiotics which are essential to its balance. This potential has not gone unnoticed in the scientific community and several projects aimed at exploring the digestive, metabolic, immune and neurological potential of gut microbiota have emerged in recent years.
One of the lines of research at the MICALIS Institute is to understand the role played by bacteria living in the digestive tract, their relationship with their host and their impact on human physiology and health. More specifically, the ChemSyBio team at the Institute is studying the natural synthesis of bioactive peptides derived from bacterial enzymes located in human gut microbiota.
As of 2013, Olivier Berteau (who manages the team’s work) has received funding from the European Union in the form of an ERC Consolidator grant. This means he can further research into the enzyme mechanisms of a highly versatile family of proteins called ‘Radical SAM’. These metalloenzymes catalyse a wide variety of complex and unique reactions. “Over the past five to ten years, their importance has been increasingly recognised in the biosynthetic pathways of natural products, including RiPPs (Ribosomally Synthesized and Post-translationally modified Peptides),” explains Olivier Berteau. RiPPs occur naturally in microbiota and have a wide range of structures and functions, including antibiotic properties in the case of some of them.
Two ERC Proof of Concept grants and a peptide with promising antimicrobial properties
In 2017, Olivier Berteau received an ERC Proof of concept grant for the D-Bug project, the aim of which was to put the team’s findings into practice. “At the start, we were trying to understand how the ‘Radical SAM’ enzymes worked. Increasingly we were also using them to find other RiPPS,” recalls the researcher.
In 2018, the team’s work was recognised with a patent application in 2020. While the D-Bug project continued, Olivier Berteau was awarded a second ERC Proof of concept grant for the ENTRAP (Engineering of natural products) project which focused on another family of RiPPs. “We know that several dozen new compounds need to be developed to address the antibiotic crisis. Depending on the application, antibiotics have different requirements in terms of properties,” adds Olivier Berteau.
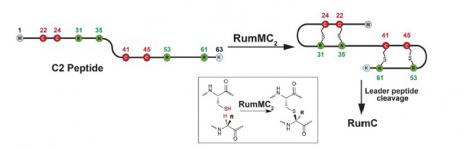
The most recent work carried out by the ChemSyBio team is to do with the enzyme mechanisms involved in the biosynthesis of Ruminococcin C (RumC), a RiPP of the sactipeptide class, produced by Ruminococcus gnavus, one of the most common bacterial strains in human gut microbiota. This sactipeptide is characterised by four intramolecular α-thioether bonds. Initiated by the putative Radical SAM enzymes, RumMC1 and RumMC2, these α-thioether bridges constrain the C1 or C2 precursor peptides into a double hairpin structure. This unique structure, which together with the removal of the signal peptide, gives RumC an active antibiotic function on a wide range of bacteria - including strains which are multi-resistant to conventional antibiotics.
A multidisciplinary approach for a better understanding of enzyme mechanisms
Biochemical and mechanistic studies of RiPP biosynthesis, including RumC, are supported by the extensive range of methods used at MICALIS. “We combine various approaches, including physicochemical, biochemical and microbiological, etc. We also benefit from the scientific community at Université Paris-Saclay in that we can collaborate with different laboratories,” points out Olivier Berteau. Thanks to this cross-disciplinary approach, the ChemSyBio team is able to further define the RumMC1 and RumMC2 enzymes, as well as the reaction intermediates.
Today, the team is continuing to investigate these mechanisms, in particular by trying to understand the role of the different groups identified on the RumMC1 and RumMC2 enzymes. A thorough understanding of these natural synthesis pathways is all the more important as RumC is thought to possess, in addition to its strong antibacterial activity, other functions of interest in terms of treatment such as anti-inflammatory properties.
References:
- Clémence Balty, Alain Guillot, Laura Fradale, Clémence Brewee, Benjamin Lefranc, Christian Herrero, Corine Sandström, Jérôme Leprince, Olivier Berteau, Alhosna Benjdia. Biosynthesis of the sactipeptide Ruminococcin C by the human microbiome: Mechanistic insights into thioether bond formation by radical SAM enzymes. J. Biol. Chem. (2020) 295(49) 16665–16677.
- Clémence Balty, Alain Guillot, Laura Fradale, Clémence Brewee, Mylène Boulay, Xavier Kubiak, Alhosna Benjdia, Olivier Berteau. Ruminococcin C, an anti-clostridial sactipeptide produced by a prominent member of the human microbiota Ruminococcus gnavus. J. Biol. Chem. (2019) 294(40) 14512–14525.
- BENJDIA, Alhosna, GUILLOT, Alain, BERTEAU, Olivier. Nouveaux peptides présentant une activité antimicrobienne et nouvelle enzyme apte à convertir un résidu à configuration l en acide aminé à configuration d dans un peptide, Brevet EP3383888 - 10/10/2018.